April 23, 2024
By Kylee Hanks and Sarah VanRyswyk
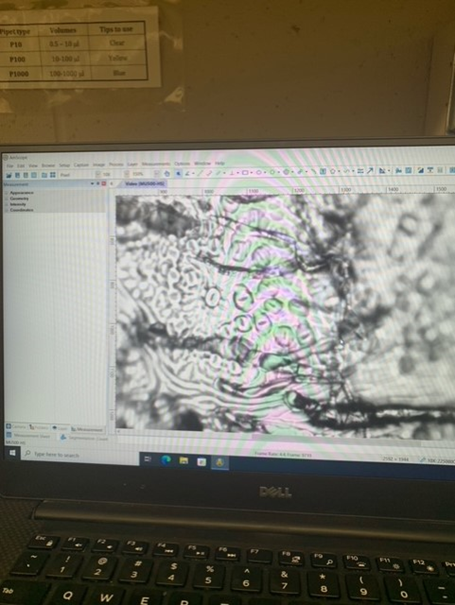
Recently in our RISEbio labs, we have been working on our research experiment. In our experiment, we were measuring the differences in osmolyte concentration in tomato plants exposed to different colored lights. Over the last few weeks, we’ve been taking osmolyte measurements from our different tomato treatment groups. There were five different pots of tomato plants under each of the three light colors. It’s been super exciting to watch our tomato plants grow and collect data to analyze, but we’ve definitely faced some large obstacles. Oftentimes, the nitrogen tank would be empty on the days we needed to take measurements. This made it hard to take consistent data every week, but we were able to get data when the tank was filled. There were other times when the osmometer would take an excessive amount of time to dry the leaf specimen. There were days we would be spending two hours in the lab to only obtain two measurements. We also ran into an issue with the leaf samples shriveling and shattering after being taken out of the liquid nitrogen.
Despite all of the complications, we were still able to get data from the tomato plants that we could then begin evaluating. Last week, we focused on evaluating our data and working on our results presentation. We ran multiple T-tests and found that our data was not significantly different. While the osmolyte concentration data was not significant, we still saw physical changes in the tomato plants under different lights.
The tomato plants that had only been exposed to white lights were by far the biggest, with blue and green plants following far behind in height and leaf count. We also saw various notable observations in the way the plants had begun to grow. All 3 treatments were very etiolated, stringy, and some had adventitious roots. This was likely due to the insufficient lights and high humidity under the pots. Overall, if we were to continue this project in the future, we may experiment with different variables including physical traits, stomata data, and leaf comparisons.