November 12, 2021
Emily Schmidtbauer and Kaela Wierenga
Introduction:
Since September 24, 2021, we have worked to insert our upstream region of the endonuclease-encoding gene into our construct pSW01-3. We were able to successfully insert our upstream region creating our new constructs pSW02-6 and pSW02-8. Knowing that both were positive, we decided to continue with one, pSW02-8.
Specific Content:
Using our ligation product, we wanted to transform it into E. coli cells to prepare for a colony PCR. We threw out our plates from last semester and created new LB/amp plates for our research. Our plates for our starting growth had very little colonies that were relatively small. From each plate, we were able to patch eight colonies on new plates. From this patched growth, we selected four colonies from each plate to continue with a colony PCR.

Figure 1: Kaela is pouring out the LB/amp plates. The plates are sterile, but she has to wave the flask over the Bunsen burner to sterilize the air around the flask before she pours them.
We took the eight colony samples and created a PCR for each one. We then ran the samples plus a control utilizing genomic DNA through a gel using electrophoresis. Four of our colonies were positive, so we chose two of the positive samples to do an inoculation. To isolate a colony, we streaked some of the original patched plate colony onto a new plate, trying to dilute the colonies to just one single isolated colony. Using this single colony, we were able to inoculate it into some LB broth with ampicillin to select for our positive colonies. Once the inoculation was complete, we extracted the plasmids from the E. coli.

Figure 2: Emily is sterilizing a glass “hockey stick” to spread our E. coli cells on our completed plate.
Since we extracted our plasmids, we ran another gel to confirm that we successfully inserted our upstream region as well as looking to see if our downstream region was still present. We also confirmed our whole upstream and downstream products were in the plasmid.
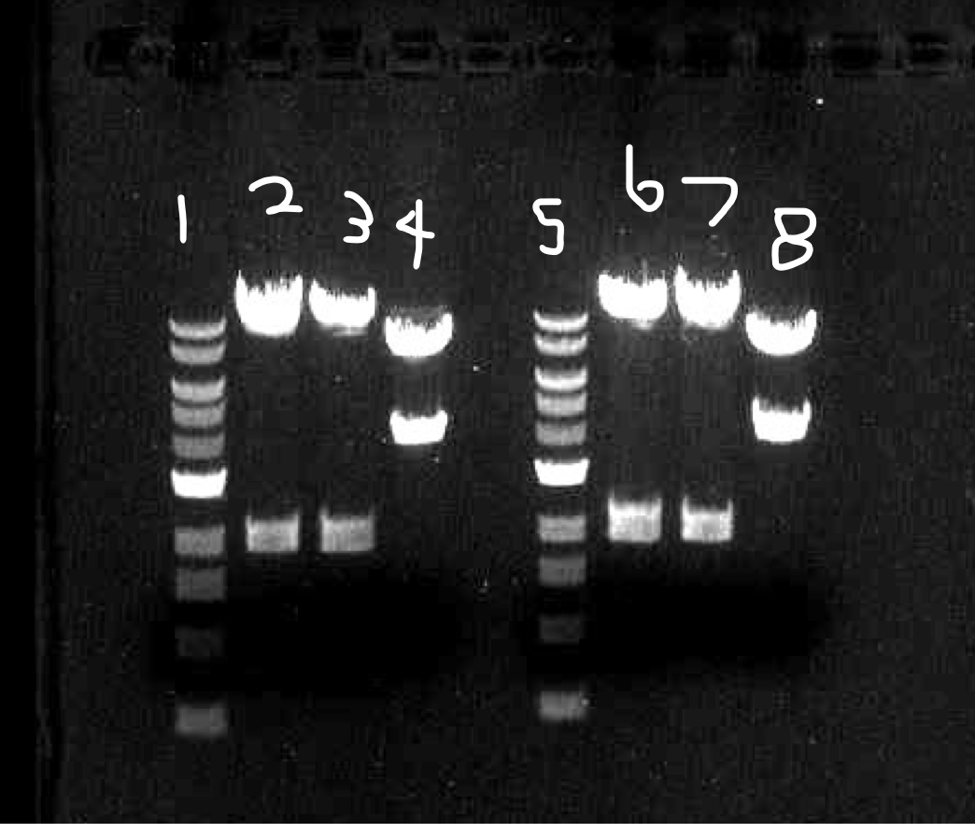
Figure 3: This is our conformation digestion gel. Columns 1 and 5 are the 1kbp DNA ladder. Columns 2 and 6 are digestion of our upstream. Columns 3 and 7 are a digestion of our downstream. Columns 4 and 8 are a digestion of our upstream and downstream.
Conclusions/Reflections:
Our research up until now has been very similar to our Spring 2021 research. The biggest difference was we used our first construct pSW01-3 in place of pYT313. We also wanted to insert our upstream region since the construct already contained our downstream region which was generated with different primers.
Now that we have confirmed that our final deletion construct pSW02-8 contains our upstream and downstream regions, we can perform a transformation. As we continue our research, we will use the E. coli S17-1 lambda pir strain to do our transformation since it has pili that are required for the conjugation process in the next step. We will utilize the product as a donor strain to conjugate our deletion construct into the pathogen Flavobacterium psychrophilum. We are looking forward to generating the final mutant lacking the endonuclease encoding gene.

