October 4, 2022
James Hawco and Jake Lohn
Introduction:
Since our last blog post, we have successfully obtained the fpsY deletion mutant from a wildtype background of F. psychrophilum. The mutant has shown a few differences (phenotypes) compared to the wildtype version. Our current goal for this semester is to reinsert the deleted gene back into our mutant and recreate the wildtype, which is called complementation. If the recreated wildtype shows identical characteristics to the normal wildtype, then we can verify our deletion was successful and the phenotypes were not caused by other factors.
Specific Content:
The work to reinsert the fpsY gene back into our mutant began with preparation of pCP11 plasmid, which can replicate in F. psychrophilum. After this was performed, we conducted a digestion for ligation with the fpsY gene to generate the plasmid for reinsertion into our F. psychrophilum mutant. These procedures were conducted in almost identical fashion as before when we deleted fpsY however, the phusion PCR process this time used new forward and reverse primers that were designed over the summer. Besides the change in primers and adjustments of time and temperature in PCR, everything else was unchanged and we were able to move on to our next steps in the reinsertion process.
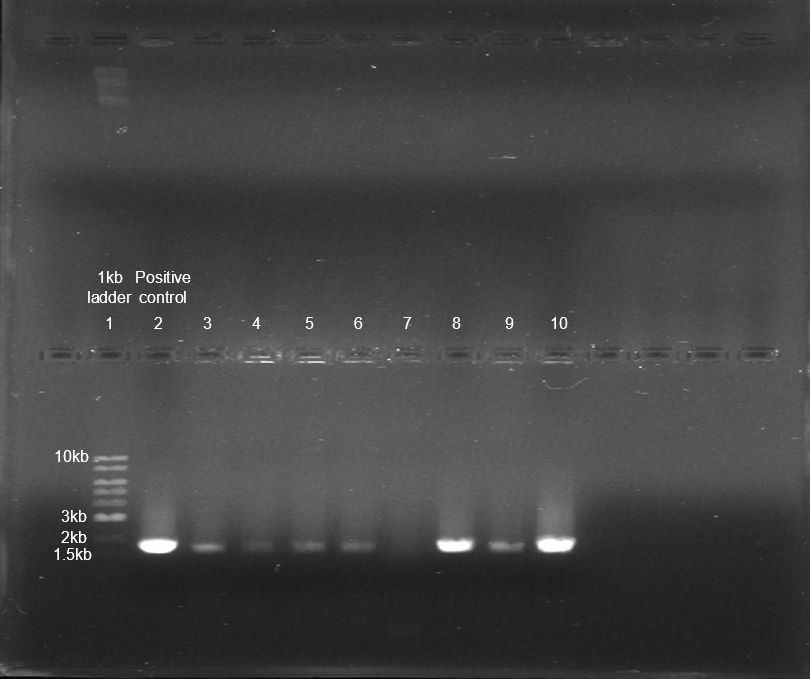
After our transformation was done properly, we began colony PCR to verify whether the colonies that grew had the expected plasmid. This process used the new primers but besides that, it was no different from our last colony PCR in the spring. After our PCR was done, we needed to run it through a gel to see which colonies had the plasmid and which ones were the most concentrated. While the PCR went without issue, the gel step had an issue arise when it was found out that the gel slid out of place from the area that it was supposed to be in which resulted in only the lower half of the gel working properly. Luckily, the lower half had the required things to verify the presence of the plasmid which are the 1kb DNA ladder, the positive control, and the colonies pl2-3 through pl2-10. Out of the colonies that did work, 7 out the 8 were positive and 2 of the colonies, plate 2 colony 8 and plate 2 colony 10, were extremely concentrated which let us move on to preparing our plasmids from these colonies for confirmation.
Conclusions/Reflections
Our work has been going successful so far but we made a few mistakes that were fixed immediately. Since we have created an E. coli strain with the insertion plasmid of the fpsY gene, the new plasmid when verified will be known as pKL03. Our future plans so far consist of confirming our plasmid as pKL03 and beginning conjugation of this plasmid into our F. psychrophilum mutant. If this is done with time to spare, we also plan on deleting the fpsY gene in Dr. Zhu’s fpsX deletion mutant to create a double deletion mutant.