4 March 2020
By: Abby Maruska and Zoe Rivard
Introduction:
Our goal for our research in the Brain and Behavior stream is to determine how our gene (SCN3B) is regulated in Anolis carolinensis lizards during the breeding and non-breeding seasons. Before we could start conducting our research on the lizards, we had to learn the basic tests. These included running a PCR in the thermocycler, creating a gel, and running it through the electrophoresis machine. Following the protocols, running these same tests, and manipulating our primers plus control primer sets (beta-actin) that were given to us; has helped us set the foundation for the rest of our research.
Research:

We selected the gene SCN3B, which is a sodium voltage-gated channel (beta subunit 3). This gene controls the generation of action potentials in neurons. Once deciding on studying this gene, we had to make the primers for them. Making primers allows us to amplify our gene through testing and determine if we are correctly sequencing it. With doing this comes a lot of trial and error. We have made numerous polymerase chain reactions (PCR) and ran the samples in a gel electrophoresis. Gel electrophoresis allows us to take a picture of our results, further allowing us to analyze it.
On one of our more recent gels, we found that our primers amplified a band at the right amplicon, but we also saw primer dimers. We concluded that these primer dimer bands are because of the concentration our primers are at. So, we choose to dilute our primer from 1µM to 0.5µM and 0.2µM. Running a PCR and gel at these dilutions showed that 0.2µM was to low and barely showed up in the gel, and that 0.5µM was going to be the best concentration of primer to test our gene at. In the results of the gel, we had primer dimers below our correct amplicon, meaning that the primers were binding to something other than our cDNA. Along with that, our water (the negative control) had DNA amplification and primer dimers.
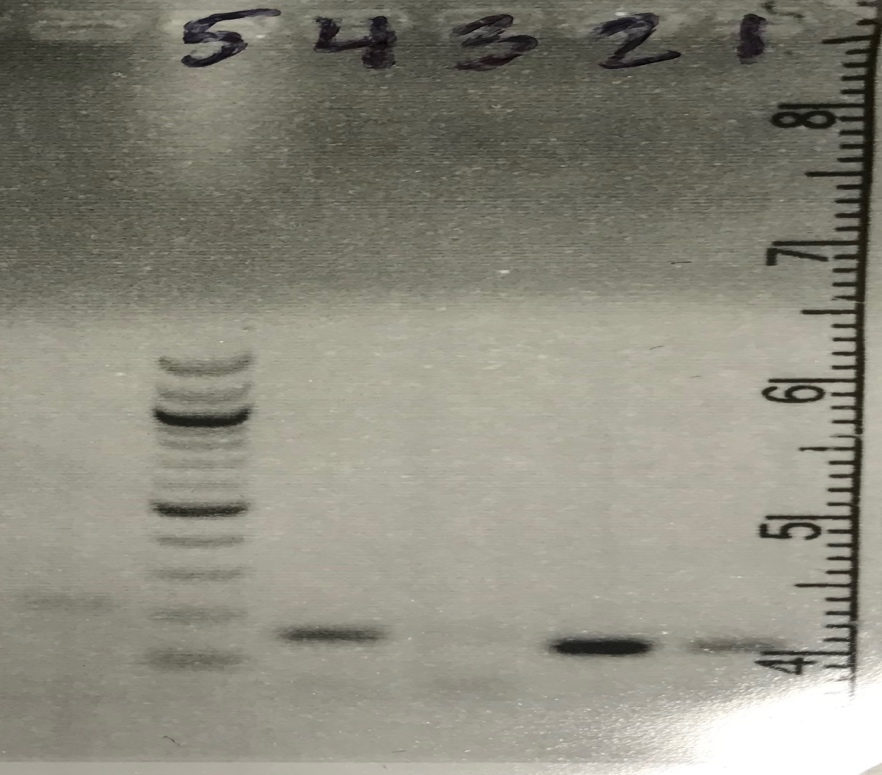
We decided to make another PCR and run it on a gel with only 0.5µM of our primer 2 and beta actin as our control, however this time with more CDNA than before. The results from making a PCR with more CDNA turned out better than the gel before (gel image to the left). There was a slight amount of primer dimers in our 0.5µM CDNA sample but everything else, water and beta actin controls, looked great. In lane 5-DNA ladder, lane 4-Pimer #2 with CDNA, lane 3-Water control, lane 2- Beta Actin control, and in lane 1- Beta Actin water control.
Conclusion:
Research is not an easy task as you endure many bumps in the road. Some of those bumps include troubleshooting our primers and running the same tests many different times. Along the way we have learned to be patient and appreciate when results look the way they are supposed to. We will continue to grow our skills learned in lab and fix our mistakes. Our future plans include running another gel for our 0.5µM diluted primer PCR and performing a PCR cleanup shortly after.

