By Kaity Shaffer and Sarah Oberstar
22 March 2024
During the last two weeks we have made some advancements in our research. We are examining how certain genes are expressed differently in breeding and non-breeding Anolis carolinensis. We have chosen to focus on two genes: KCNJ4 and NR1D2. A protein that acts as a channel inside cells, especially in the heart and brain, is encoded by the KCNJ4 gene. On the other hand, the NR1D2 gene affects metabolic processes and controls the body’s biological circadian rhythm.
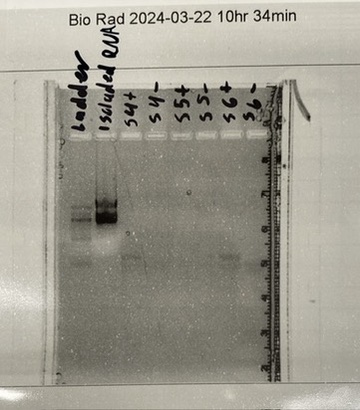
After many PCRs and gel electrophoreses, two primer sets for NR1D2 were good enough to clean up and send into sequencing. The primers showed a dark and correct band size in the gel images. However, the KCNJ4 gene was not showing up correctly. Only primer dimers were showing up in the gel meaning the DNA was degraded and not being expressed. We had to redesign three new primer sets for the KCNJ4 gene.
With two good looking primer sets, we moved on to the DNA sequencing. To do this we first had to do a PCR clean up, meaning we purified the DNA. Impurities like excess nucleotides and primers are removed to produce pure DNA samples. We then sent our two NR1D2 samples in to be sequenced using Sanger sequencing. Once we got our results back, we compared our sequence to the target sequence using 4Peak Software. Luckily, we correctly cleaned the PCR the first time. Our sequence came back with a 99% match. This indicates that the primers we have are successful and are amplifying the correct sequence.
The next week we practiced RNA isolation. In this step we would usually use Anolis carolinensis brain, but we were just practicing so we used their livers instead. We first homogenized our liver sample. The homogenizer was a very cool tool to work with. We then removed cell debris and proteins using chloroform. This was also fun to work with because we got to wear lab coats. Lastly, we eluted our solutions. To do this we got to use a fancy centrifuge.
Overall, the last two weeks have gone well. We have learned a lot of new things and have moved on to new protocols. By practicing RNA isolation, we are set up to do well when working on our own samples. The success of our PCR clean up and sequencing is a reflection on how far we’ve come while working in the lab. We used to have to do a protocol a couple times before we got it right. Our next steps are testing our new primer sets for the KCNJ4 gene. Once the primers are amplifying correctly, we can move forward with the same process we put the NR1D2 gene through. We can also move on to isolating Anolis carolinensis brain RNA now that we are done practicing.

