22 April 2023
By: Brenden Bauman and Rebecca Fasching
This semester we are continuing our work on studying the brain and behavior of the Green Anole lizards (Anolis Carolinensis). Our current focus on this project is the isolation of RNA from the brains of Green Anole lizards. This RNA is to be used in the future for the synthesis of cDNA which is to be used to continue studying specific genes, which for our group would be STAB2. We are currently through three of our five assigned brain samples for RNA isolations.
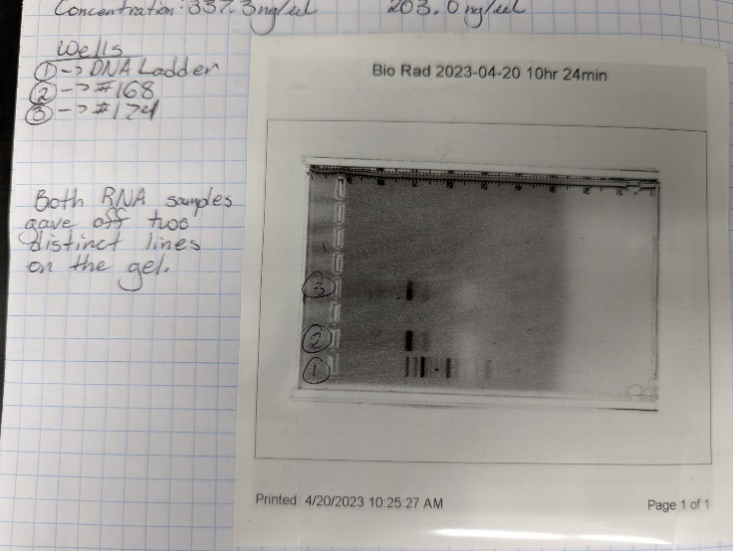
In the last few weeks, we have been working on our brain RNA isolations of the Green Anole Lizard. We began last week’s lab by isolating one of our brain samples RNA from the Green Anole Lizard labeled #160. This was a slight change from the previous week as we had been using liver samples to practice RNA isolation and with the introduction of brains to run an RNA isolation, we had to add different steps to our protocol. That included creating a mixture by adding DNase 1 and Buffer RDD together. Which was then later added to our samples through our columns. That day we also used the nano dropper to get the A260/A280 and the concentration (ng/µl) of the brain RNA. The next lab we spent time working on a gel electrophoresis of the RNA to confirm that the RNA was not degraded. The week after that we did the same thing, but with two Green Anole brain samples. Once again collecting the A260/A280, concentration (ng/µl), and running a gel electrophoresis of our RNA.
The results of our troubleshooting tests showed that our RNA isolations turned out well. Our first brain sample, number 160, came back with a 260/280 ratio of 2.14, a concentration of 304.6 ng/µl, and two bands on the gel electrophoresis. Number 168 came back with a 260/280 ratio of 2.14, a concentration of 337.3 ng/µl, and two distinct bands on the gel. Number 174 had a 260/280 ratio of 2.11, a concentration of 203 ng/µl, and two bands on the gel. Overall, every 260/280 ratio was around 2 which is the desired result from that test. Each sample showed two distinct bands on their gels which means that the RNA was not degraded during the isolation process. So far, all of our isolations have been a success.
In the end our labs are going well, we have, for the most part, been very effective and efficient in our work. While learning how to properly run an RNA isolation, how to properly use a nano dropper and furthering our ability to run a gel electrophoresis. We feel that our results show this with all our RNA A260/A280 being around 2.0, our concentrations (ng/µl) being very close between samples, and all our gel images showing a very clear two bands per RNA sample. Still in some instances we find that one lab partner is taking more of the blunt of the work. However, this gives us the opportunity to learn from our mistakes and try to evenly distribute work better. Our pipetting can still be improved as well; however, we have learned more effective ways to pipet and are learning different tactics each day. Overall, we continue to perfect our skills in lab while furthering our research of the Green Anole Lizard and the STAB 2 gene.