April 16, 2021
Emily Schmidtbauer and Kaela Wierenga
Introduction:
In this blog post, we will further analyze our endonuclease gene and look at what our next steps have been in our research since April 1, 2021. We are still looking at the Flavobacterium psychrophilum bacteria with our endonuclease gene which catalyzes the cutting of nucleic acids.
Specific Content:
When conducting research on our endonuclease gene, little was found. We were able to find information on what an endonuclease is and its function, but little is known about how it relates to the virulence of Flavobacterium psychrophilum bacteria. The endonuclease gene is not a restriction endonuclease meaning it will randomly cut sections in a nucleic acid sequence. Because of this randomness, we hypothesized two ideas. The first idea was the bacteria could digest the DNA nucleic acids from the fish cells to smaller fragments and use these as nutrients. The second idea was the bacteria would destroy the host DNA, leaving the nucleic acid incomplete, meaning the host cell is not able to replicate the nucleic acid sequence, resulting in no cell replication and a depletion in cell count and organism growth.

Figure 1. Emily and Kaela work to transfer each of the five E. coli samplesinto a microcentrifuge tube.
After transformation of the DNA ligation product, on our E. coli plates, we had a significant amount of growth, so we were good to continue with our research. We each took one fresh LB agar plate and split the plate into ten sections. Using a sterile pipet tip, we picked up one colony on our growth plates and streaked it onto our new plate in one section. After disposing the pipet tip, we repeated the process until all our ten sections had a colony spread on it, having twenty sections total between our two plates. We then incubated our new plates for twenty-four hours at 37oC. The result was quite a bit of growth, so we picked out five sections from each plate to conduct a PCR test on.
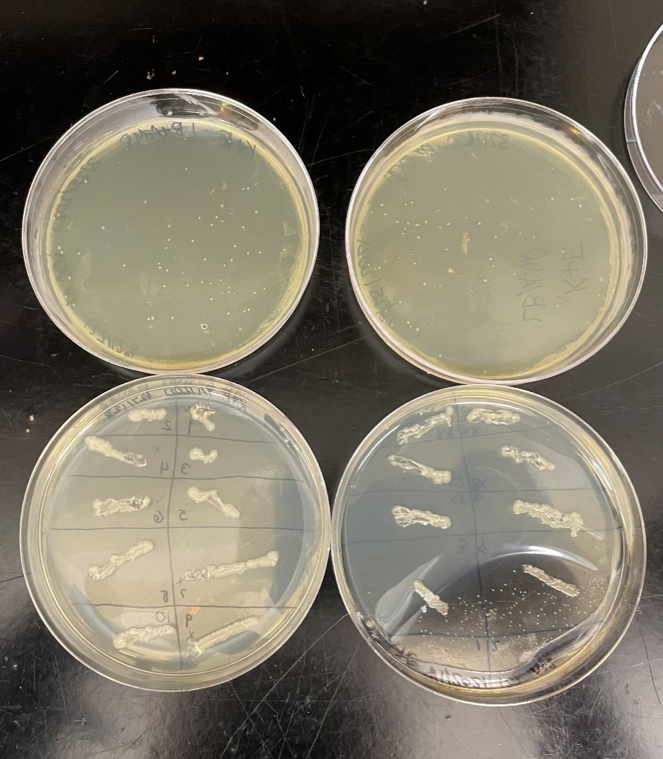
Figure 2. The top row is our original agar plates with E coli growing after transformation. The bottom row is the ten colonies we each streaked onto a plate.
The purpose of the PCR and running a gel was to know which colonies took our ligation product. In our gel, we included a 1 kb ladder, control sample with genomic DNA as the PCR template, and our ten samples. All our samples were PCR positive and the expected DNA bands appeared on our gel, but only about half were very bright. We continued with two colonies that we think were the brightest on our gel and streaked each on a new LB agar plate for isolation of pure cultures. We incubated the plates at 37oC for roughly twenty-four hours. After we saw colony growth, we did an inoculation into a test tube with five milliliters of LB broth and five microliters ampicillin. Since our plasmid is ampicillin resistant, only the cells with our plasmid would grow. We shook our sample at 175 rpm in an incubator at 37oC for twenty-four hours. After this was completed, we transferred 1.5 milliliters from the incubated test tubes into a microcentrifuge tube and completed the plasmid extraction process.
Conclusions/Reflections:
Little is known about the endonuclease gene on the virulence of Flavobacterium psychrophilum, but we believe our research can show if they are related. We completed the screening process of our transformed E. coli cells and found several colonies that took our correct ligation product. We continued with two colonies and extracted the plasmids. As we are soon concluding with the downstream region of our endonuclease, we are going to trouble shoot the upstream region and start over our whole process, but with new primers.

