13 September 2019
By Nicole Michealson and Marc Nganteh
Introduction:
In the spring semester, we ended off our experiments by doing RNA extractions and multiple PCR cleanups. We use the brain of Anolis carolinensis (green anole lizards) to obtain mRNA at each time point to see how the genes are expressed. We are trying to see if gene expression patterns are different during the breeding versus non-breeding seasons of the green anole lizard. To begin this process, we remove the cell membrane to isolate the mRNA by using a homogenizer, which shreds the tissues. Then we put it in a column and use chloroform to remove cell debris and proteins. To make sure there is no gDNA, we use DNase to break down any gDNA that could be in the tissue, and then we wash it down with buffers to ensure there is only mRNA left. The final step is to check the RNA integrity by using the nanodrop to check concentration and RNA purity, along with doing a gel electrophoresis to assess if two bands appear.
In the spring semester, we sent in multiple PCR cleanups that had not amplified our TSHb gene correctly. After the semester our teaching assistant, Hyejoo, tried doing a PCR cleanup on our gene, and it still did not amplify our gene correctly. A PCR cleanup is used to remove enzymes, nucleotides, primers, and buffer components. The only thing that should be left is our TSHb gene, or the amplicon. The TSHb gene has 84 base pairs with a specific code, which is what should be amplified.
Research:

We started the fall semester and were asked to run PCR on our respective genes to refresh our minds of what we were doing in the previous semester. We ran our PCR by preparing a 2x master mix at 61.8 ℃ in the thermocycler, which should be amplifying our gene. For the PCR, we had two PCR tubes of our TSHb gene, so we could use one PCR tube to do the PCR clean up with Dr. Cohen. When running our gel electrophoresis to visualize the results of the PCR, nothing appeared on the gel image. We think we could have made an error putting gel loading dye in our gel instead of the gel stain (Bullseye). Otherwise, there could have been a problem with the filter on the gel imaging machine, or we put in the gel stain when the gel was too hot.

The following week had to then do another PCR and this time we also created a 2x master mix for Beta-Actin to be used as a positive control, with cDNA in one PCR tube and water control in the other. We also created 2x Master Mix for our TSHb so that we could run our PCR cleanup with Dr. Cohen using a better, newly purchased kit. This time the gel electrophoresis and imaging showed the DNA ladder, the TSHb gene, and both Beta-Actins controls. The band for our gene showed up around 84 base pairs, which is the amount it is supposed to be at. We also saw a light band for the negative beta control (the one which had water control). This could mean that our PCR H2O was contaminated. We went on to do the PCR cleanup with Dr. Cohen. Our past PCR cleanups could have failed because our amplicon length (84 base pairs) was too small and got washed out during centrifugation.
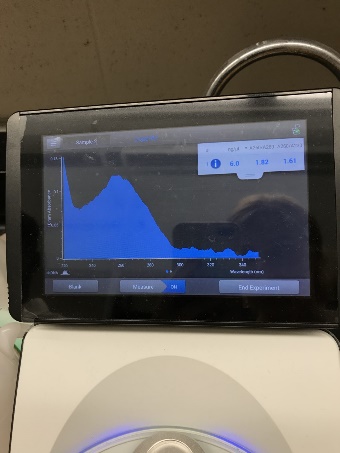
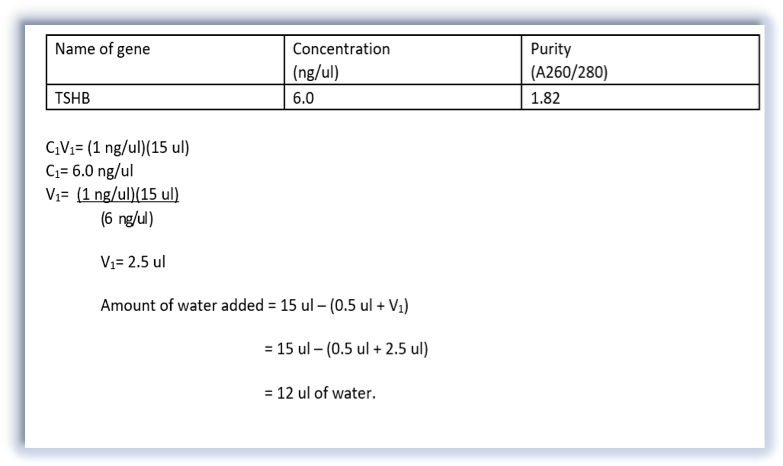
During the PCR cleanup, membrane binding solution and membrane wash solution is added to the PCR sample and centrifuged, to remove any proteins or enzymes that were in the sample. Our desired DNA was then separated from other contaminants by adding Nuclease-Free water and centrifuging for 1 minute. We then checked the concentration and purity of the DNA using a nanodrop, added the appropriate amount water and forward primer to our sample and stored the purified DNA in the freezer at -20℃. This sample was later sent out for Sanger sequencing. All this information was sent to our lab instructor, Dr. Cohen, who sent it out for sequencing later. This process determines if the DNA sequence that was amplified during PCR was for our TSHb gene. This is a kind of PCR where ddNTPs (which have fluorescent dyes) are added to the template. If the result comes out positive, we would analyze, export, and blast our sequence to the anole genome.
Reflection:
As previously stated, we performed a PCR this fall semester to refresh our minds. We were confident in what we were doing because performing PCR is one of the lab processes we performed before in the previous semester. The PCR failed on the first trial, but we figured out our mistakes to get adequate results from the PCR and gel electrophoresis. Our gel imaging results showed a thick band at the correct base pair size for our TSHb gene. I would say the most tedious task was performing the PCR cleanup since we used a kit that we’ve never used before, but Dr. Cohen helped us through the whole process. The higher quality PCR cleanup kit is a new strategy we are using for our PCR cleanup to show correct results, and hopefully will be a stepping stone in our goal to see if the TSHb gene of thegreen anole lizards is differentially expressed for males and females in the breeding versus non-breeding seasons.
Future Research:
Next week in the lab, our group will begin doing cDNA synthesis on the mRNA we collected from the brains of the green anole lizard. This is the synthesis of DNA from an RNA template via reverse transcription to produce cDNA. This happens by using dT primers containing thymine which is complementary to adenine on the poly-A-tail, which is the 3’ end of the mRNA that contains a lot of adenine. For the cDNA synthesis protocol, the first step will be to use a heat block to denature the mRNA. Then the mRNA is placed in an incubation chamber to synthesize cDNA. By the end of the protocol, we will store the cDNA -20℃ freezer. We have 8 samples to do the cDNA synthesis on. Once all groups in the Brain and Behavior stream have finished, we will all run PCR’s to see if they show the bands they are supposed to on the gel electrophoresis and imaging.
References:
Cohen, R. (2019). Lecture 4 – RNA Extractions. PowerPoint presented in the RISEbio Brain and Behavior Class at Minnesota State University – Mankato.
Cohen, R. (2019). Lecture 3 – PCR Troubleshooting. PowerPoint presented in the RISEbio Brain and Behavior Class at Minnesota State University – Mankato.
Cohen, R. (2019). Lecture 8 – cDNA synthesis. PowerPoint presented in the RISEbio Research at Minnesota State University – Mankato.
New England Biolabs Inc. (2019). PCR & Reaction Cleanup. Retrieved by: https://www.neb.com/applications/dna-amplification-pcr-and-qpcr/pcr-and-reaction-cleanup

