September 17th, 2019
By Kaela Goodman, Meaghan Keohane, and Ramsey Pankratz
Introduction
The goal of our research is to predict miRNA regulation of the APOBEC3A and APOBEC3B genes. We started by selecting specific miRNA to work with and have since cloned and sequenced them. Currently, we need to transfer our miRNA from the cloning vector to an expression vector. Originally, we needed a cloning vector to multiply, or clone, our miRNA. However, now that our miRNA have been cloned, we need to proceed to the next phase of our research; we need to predict whether or not our miRNA can regulate APOBEC3A and/or APOBEC3B. In order to do that, our miRNA need to be in an expression vector. Thus, we must now transfer our miRNA from the cloning vector, pMiniT 2.0, to an expression vector, pcDNA3.1+.

Figure 1. Meaghan carefully cutting out each of the target bands
Our Process
This week we resumed work with two of our microRNAs, hsa-miR-4522 and hsa-miR-890, by preparing both our microRNA
and the pcDNA3.1+ vector to be ligated.
We
began by performing restriction digestion on our microRNA; a process which would cut it out of
the cloning pMiniT2.0 plasmid using the
restriction enzyme EcoRI. In order
to accomplish this, we
simply combined EcoRI and
our microRNA (either 4522 or 890), along with other
components, and incubated them for an hour. Next, we performed gel electrophoresis to visually check if
we were able to successfully remove each
microRNA from pMiniT2.0. We
were able to see that our process was successful for both 4522
and 890 due to the
presence of two bands, one for the cut out microRNA fragment and another for
the
rest of the plasmid.
To prepare pcDNA3.1+, the expression vector, to be ligated, we performed both restriction digest and phosphatase treatment on the vector. The restriction digest portion of this process directly follows what we for the microRNA. However, after incubation, we performed the phosphatase treatment to pcDNA3.1+. This step cleaved off the phosphate groups on the ends of the cut vector. Thus, rendering it unable to ligate back together with itself. We were then able to visualize the product of these steps via gel electrophoresis. Only one band was present and at the right base pair length after visualization which indicated that we successfully performed the restriction digest and phosphatase treatment.
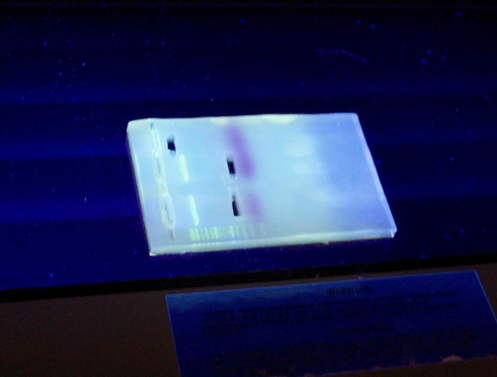
Our final step of this week was to physically cut out our microRNA
and pcDNA3.1+ out of the electrophoresis gel. By
placing our gel with
our samples on a UV light box, we were able to see where each of
our DNA fragments were.
Thus, Meaghan was able to
physically cut out each
target band from the gel, the bottom band, containing the microRNA
fragment, and the
pcDNA3.1+ band, creating a
total of three gel plugs.
Future Steps
As we move forward with our three gel plugs, we will melt each plug into liquid form and purify each sample individually. After gel purification has been completed, we will test each miRNA sample and the pcDNA 3.1+ vector on the nanodrop. If the nanodrop identifies the samples as deoxyribonucleic acid and the concentrations (ng/ul) are high enough, we will move on to ligation and transformation.
In the ligation step, we will combine the vector with each insert. After everything has been combined, we will use a varying array of temperatures to perform transformation, which will cause the bacteria to take up the plasmid. The final step to transformation will be to plate our samples and allow them to grow overnight.

