13 FEBRUARY 2019
By Anne-Marie Akinpelu & Platung Chang
Our study focuses on how the immune system’s response to viruses can also damage our healthy cells and, over time, cause cancer. The research that we will be working on over several weeks, will demonstrate how our immune system damages cells within us as it fights the HIV virus. As it is also able to fight other viruses, the immune system deliberately damages HIV by embedding dangerous mutations into its genetic code. At times, our immune system may have difficulty recognizing help from hindrance and can generate those mutations in our DNA. Eventually as time goes on, these mutations accumulate, causing cancer.
My partner and I are working to investigate questions that are centered around cancer and immunity. We use molecular biology to study inherent immunity to viral infections, host-pathogen interactions, and possible abnormalities to infection as well as autoimmune diseases and cancer. We conduct labs to explore ways to prompt our immune system in the right direction, so it is able to fight viruses and not our bodies. We are especially interested in APOBEC3A, an inherent immune protein found in humans, primates, and other mammals. It is a DNA cytidine deaminase with antiviral effects. This can lead to a positive effect if the DNA substrate is a virus, such as HIV, but could lead to negative effects, if the DNA substrate were our own, genetic DNA. We are interested in figuring out how it is controlled.
During the first week of our research we identified microRNAs that putatively targeted APOBEC3A and/or APOBEC3B, by visiting miRDB; an online microRNA target predication database. While on the website we were able to view a list of microRNAs predicted to bind APOBEC3A and/ or APOBEC3B. A target score was attached to each microRNA, with 100 as the highest possible score. We then saved our information and compiled our findings into a table.
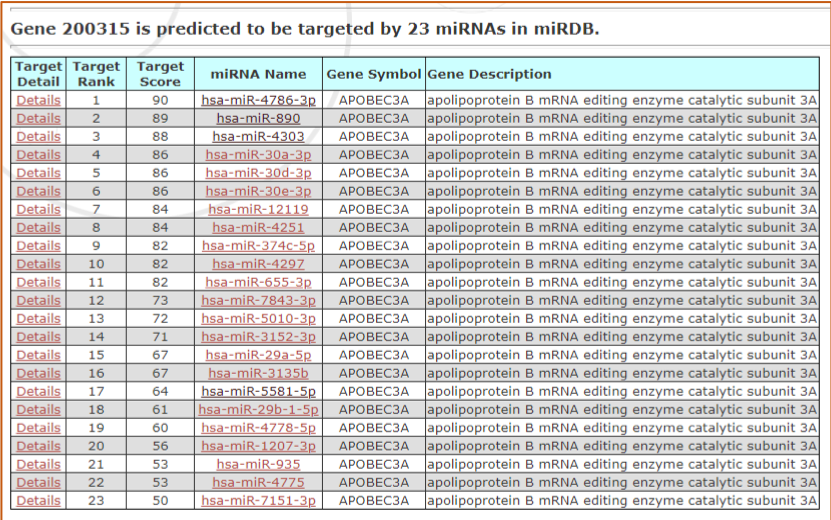
Figure.1 The list of microRNAs predicted to bind APOBEC3A in miRDB
During the second week, we were able to choose two unique microRNAs of our choice, from the list we compiled into our table from the previous week. My partner and I chose the microRNAs hsa-miR-4314 and hsa-miR-30a-39, considering they had high target scores. The higher the target score the more certainty we have in our prediction. We became aware that a predicted target, with a prediction score greater than 80 was most likely to be real.
We worked to retrieve the sequences for our selected microRNAs by Downloading and installing ApE; a plasmid sequence editor, onto the computer. We then visited GenBank; NIH genetic sequence database, an annotated collection of all publicly available DNA sequences. In the search box, we typed in one of the names of our selected miRNA. We were then able to see a list of hits. Considering miRNAs are less than 100 bp, we were able to select the one that was most suitable. The accession number and other important information was recorded into our lab notebooks.
Figure 2. NIH genetic sequence database, viewing miRNA hsa-miR-4314
In order for the miRNAs to “work” in an overexpression system, we needed more genetic information. We visited the UCSC Genome Browser; an interactive website offering access to genome sequence data from a variety of vertebrate and invertebrate species and major model organisms, integrated with a large collection of aligned annotations.
In the search box, the name of our miRNA was typed in. We could then observe that region of the human genome. After copying and pasting our DNA sequence, into the ApE file we selected and copied the miRNA sequence that was retrieved from GenBank. Additional steps were followed and was useful in helping us find a primer. Our final step was to design primers for our selected microRNAs. Designing oligonucleotide primers is an important step for successful molecular biology experiments that require the use of PCR. To design our primers, we visited Primer3Plus, and followed additional steps to create our forward and reverse primers so that we can use PCR to amplify our miRNAs in a subsequent lab.

