By: Nikita Damyan, Losel Baro, Zhelah Guzeh
Introduction:
We have come to the end of our group’s part of the research and process to create a deletion mutant of the adhesin and motility gene, sprB, in the fish pathogen Flavobacterium psychrophilum. Our last blog involved streaking on erythromycin and sucrose plates in a process of first and second recombination. During this procedure, we confirmed that we had mutant colonies/cells which had lost the sprB gene during recombination via a colony PCR.
Specific Content:
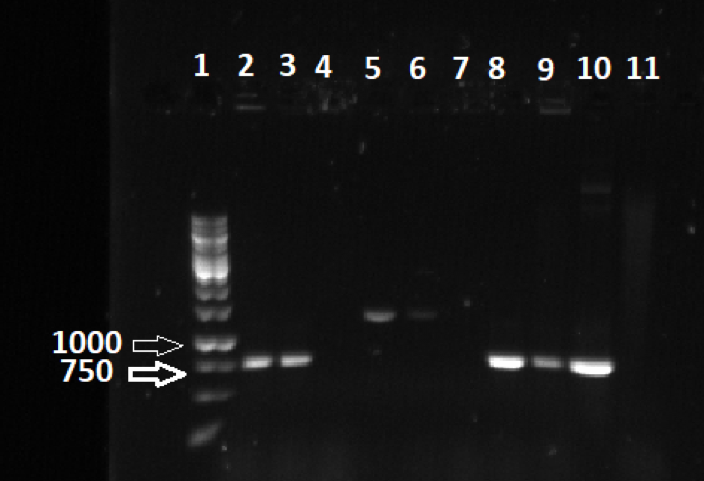
To confirm that we did have a mutant, we performed a PCR and a gel electrophoresis. At this point in our experiment, we had successfully created a mutant F. psychrophilum bacteria lacking the sprB gene. The process of PCR and performing a colony PCR gel was a little rough, as initially, the PCR gel needed to be done a total of three times, all of which did not yield the results we were looking for. It was most likely due to an error in the PCR procedure. After attempts and failures of running a desirable gel, the PCR was redone, and two more gels were run. After the failure of the first gel, a new master mix was created and a second gel was run utilizing both master mixes, both the old and the new one. This gel finally yielded the results we were looking for. It is important to note that sprB is a very large gene, and the total size of the wild type is 10526 base pairs long. Because of this, all of the wells that show bands are mutants.
With the creation of sprB mutants, we were finally able to begin testing whether the deletion of sprB inhibited the motility of F. psychrophilum. We ran a very simple test of growing a mutant colony and a wild type colony on 5% TYES agar plates and incubated them for 10 days. 5% TYES agar was used to test the colony-spreading ability of our mutant and wild type. Since 5% TYES agar plates have a diluted amount of nutrients compared to a normal TYES plate. The colonies are forced to spread to continue taking in what few nutrients are present necessary for both continued growth and life.
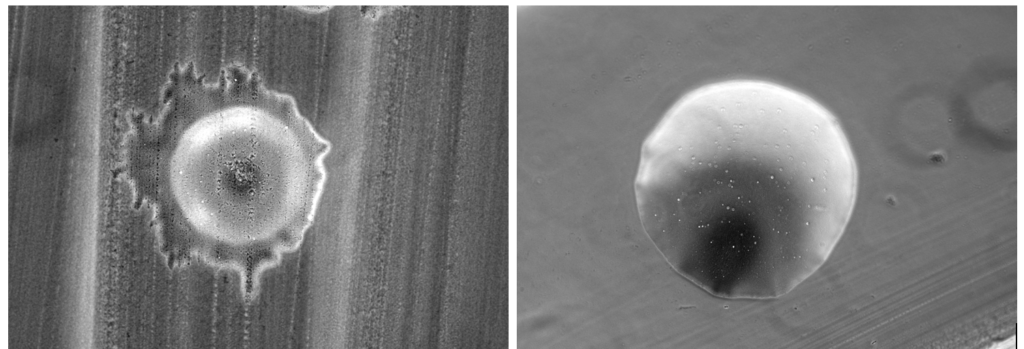
This phenotype was what we have been trying to achieve throughout the entire research stream as it showcases and supports our expectation that deleting sprB reduces the motility of F. psychrophilum. On top of these experiments, we also did a side experiment that involved inserting sprB back into the mutant for experimentation purposes called complementation. One of which was to test whether the bacteria would be motile again now that sprB was inserted into it again. We did no further tests on the complemented strain, and as such, no further information can be given about it.
Conclusions/Reflections:
Definitely one of the rougher experiments we’ve gone through due to all of the problems we had with the PCR and gel. Other than that, we agree that these last few experiments have been some of the most rewarding experiments as it shows the fruit of all of the work we’ve put into this. The future plans for this experiment involve further testing and comparing the wildtype, mutant, and complemented strains on their abilities to cause diseases in rainbow trout. The sprB mutant can be a potential vaccine if it fails to cause diseases. Huge thanks to Dr. Zhu for being here with us along this road and always being there to go out of his way to help us whenever we needed it. Big thanks to our past mentors Seada Sloboda, Emily Schmidtbauer, and Nick Shankey for watching over us and helping us during the experiments. Special thanks to the other members of this research stream Jake Lohn and James Hawco for working with us and supporting each other. Thanks to Dr. Sharlin, the leading RISEbio professor and the person who got us ready for these streams, and the NSF grant for making RISEbio possible.

