October 29th, 2019
By Ramsey Pankratz, Kaela Goodman, and Meaghan Keohane
Introduction
To date, we have amplified our microRNA, cloned it using a cloning vector, and are now transitioning to test its regulatory effect by using an expression vector. At each point, however, we must identify that the sequence we are working with is still our chosen miRNA. To do this, we first insert our vector into a bacteria and grow it on petri dishes containing an antibiotic. Since the vectors we use contain an antibiotic resistance, only cells with our vector will grow. Then we process the cells that grow and determine if their copy of the vector contains an insert. We call this screening. We screen our clones so as to not waste time and money on sequencing samples that have no chance of being what we are looking for. In the past, when we cloned our sequence using the pMiniT2.0 Cloning Vector, we screened our clones via restriction digest. Put simply, we removed our vector from the cells that grew, and used an enzyme to cut out any sequences that were added to the vector. Then we visualized the results to determine if indeed anything was “cut out,” and determine the likelihood that it is our chosen miRNA.
This week, however, we are screening samples in a different way: by Colony PCR. This method of screening functions the same as our initial amplification of our miRNA. However, this time we will use our pcDNA3.1 vector as the template instead of HeLa DNA. If the pcDNA3.1 vector contains our miRNA, then it will be amplified and visualized, if it doesn’t, then…well, then we do it again!

Our Process
To begin the PCR screening, we began by creating a master mix for our PCR reactions. In order to achieve one part of the screening aspect for this protocol, we added the microRNA’s specific reverse primer to their respective PCR reactions. This allows only our microRNA sequence to be amplified during PCR due to the primer only being able to bind to and amplify our microRNA sequence. We also used a general T7 forward primer instead of our microRNA’s forward primers, which would help to prevent amplification of any microRNA sequences that were inserted backwards into the vector.
Once we had our master mix ready to go and aliquoted, we picked colonies off of our previously made LB-ampicillin plates and added them to their respective tubes. These plates contained cells that theoretically contained each of our microRNA sequence in the pcDNA3.1(+) vector. Additionally, we created replica plates of each picked colony. The replica plates were created so if we got positive results from the colony PCR, we could ensure we moved on with that exact vector.
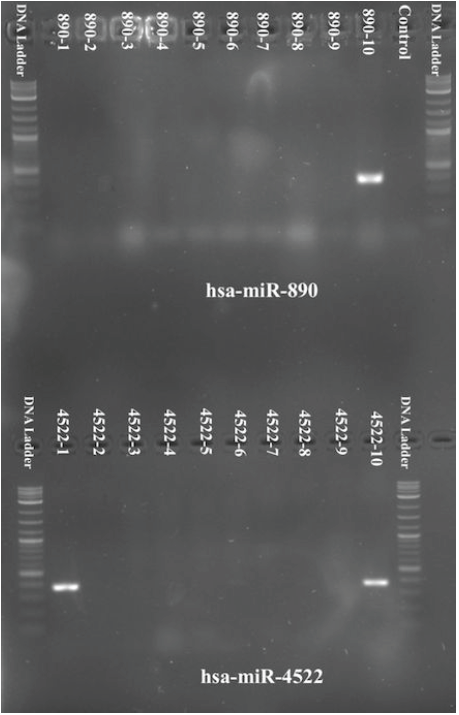
Finally, after PCR was run, we visualized the results via gel electrophoresis. For hsa-miR-4522, two of the samples showed positive results for its microRNA sequence. One sample for hsa-miR-890 showed positive results as well.
Future Steps
From our lab experiences this week, we have utilized a new technique to screen clones using PCR to check if our miRNA was inserted into the vector. We also developed skills for making replica plates so we could isolate and clone individual colonies. Since we attained positive results for two of our samples for has-miR-4522 and one of our samples for has-miR-890, we will grow each sample overnight in order to increase the concentration of DNA. After the concentration of DNA has been increased, we will purify each sample, and send them out for sequencing. The sequencing results will tell us whether or not we successfully inserted our miRNA into the pcDNA3.1(+) vector.

