April 9th, 2022
James Hawco and Jake Lohn
Introduction:
Since our last blog post, we have conducted DNA ligation, transformation, colony PCR, and confirmation for the upstream region of our target gene. After confirmation of our upstream region, we have begun progress on our downstream region. As of the last lab, we have performed colony PCR and all the other experiments for the downstream region.
Specific Content:
In the lab we did after our previous blog post, we performed ligation which seemed to go well but, the colonies we had were very few. We had to redo this process and realized we used the wrong product for it which resulted in the failure of the colony growth. After we did the process again the colonies grew properly, and we moved onto replating of specific colonies. We selected 20 of the isolated colonies to patch on a separate LB agar plate to use for PCR. After the colonies grew to a sufficient size, we had 10 selected colonies undergo PCR for testing.
The PCR process went without issue and the products were put through gel electrophoresis to test if they were positive or not. Our results showed 7 out of the 10 colonies were positive but, during the PCR process, the labels on colonies 6 (well 8) and 8 (well 10) were smudged so we did not know which colony was the positive one therefore we ignored the positive colony in well 10. Well 1 contained our 1kb DNA ladder, well 2 had our positive control, and wells 3-12 contained our colonies in increasing numerical order. Our positive colonies were 2, 3, 4, 5, 8 (potentially colony 6), 9, and 10. We proceeded to take colonies 2 and 9, due to them having the highest concentration, and proceeded to streak them to have new colonies grow from individual cells. We then proceeded to take the isolated colonies and inoculated them in a test tube to perform plasmid confirmation to see if they were positive or not.
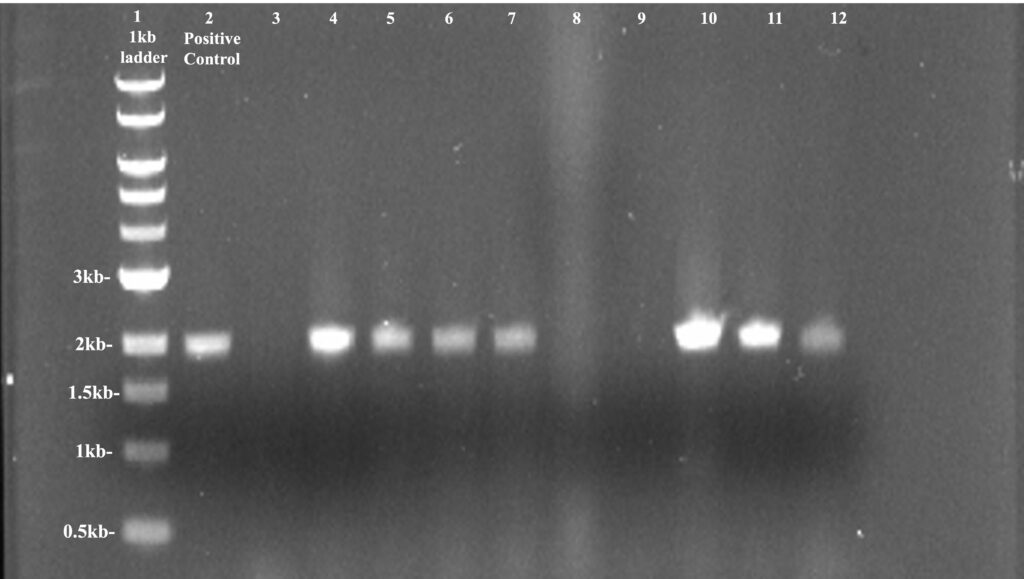
Our plasmid confirmation confirmed that both colonies had the plasmid we wanted, so we kept colony 9 which had a higher concentration to use for future projects that now have the plasmid pKL01 instead of pYT313 due to the changes we have made to it. Since we finished on the upstream region with ample time, we have begun to work on the downstream region. We began by putting the downstream region through digestion, ligation, and transformation which had potentially resulted in bacteria containing the downstream region. We performed PCR on 10 selected colonies however, when they were put through a gel the results came back negative for every colony. Due to this issue, we will be conducting PCR on the other 10 colonies to see if there are any positives and if there aren’t any, we will be redoing the ligation.
Conclusions/Reflections: For the upstream region, we had positive colonies and successfully created a new plasmid pKL01. For the downstream region, on the other hand, our colony PCR was not successful. We will be conducting another PCR on the rest of the colonies in the near future and will potentially see positive results before the end of this semester.


